March 29th, 2023
Author: Khanak Jorwal
Editor: Dr. Jitendra Kumar Sinha
Everything begins with a thought-definitive or distending. Every thought begins with a connection-visible to the eye or out of sight, in your mind. As you read this, your brain shall attempt to join the dots (neurons saying ‘hi’ to each other) and make sense of each statement that ends with a full stop (neurons become ‘friends’).
“In an extreme view, the world can only be seen as only connections, nothing else. I liked the idea that a piece of information really defined only what it is related to, and how it is related. There really is little else to mean. The structure is everything. There are billions of neurons in our brains, but what are neurons? Just cells. The brain has no knowledge until connections are made between neurons. All that we know, all that we are, comes from the way our neurons are connected.” -Tim Berners Lee ‘TimBL,’ inventor of the World Wide Web
I shall now begin to speak of insects, for some of you this word might create an image of a bright colored, little winged creature that shines through the dark or perhaps of a blood sucking monster that doesn’t let you sleep at night (so you just experienced the linking of neurons to form an image in your head based off your memory, yes, ‘see’ what I did there).
Well, as for neuroscientists around the globe, an insect’s brain is of great interest to chart the neurons in order “to trace the nervous filaments through the substance of the brain, to see which way they pass and which way they end.” One such neoteric study left the world of science (and insects too, maybe) amidst a torpedo of immense future revelations about what we could know about the brain and the underlying neural mechanism of a thought.
The International team led by John Hopkins University and University of Cambridge produced a magnificently detailed diagram tracing every neural connection in the brain of larval Drosophila melanogaster, commonly known as fruit fly. (Fig.1)
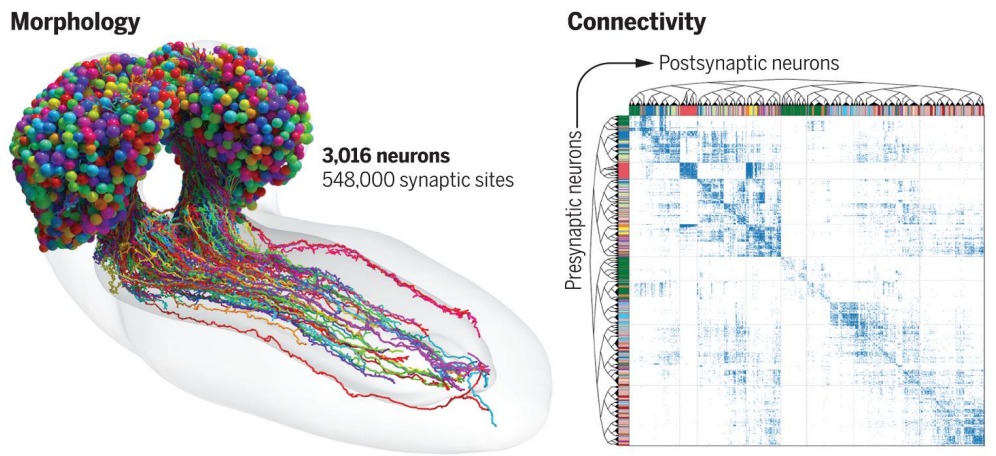
Source: https://www.science.org/doi/10.1126/science.add9330
It is quite a fascinating fete that when we are looking at something like an insect, they are doing so much, such as bees building an army to sting the person who disturbed their hive or flies forming a swamp over your scrumptious food. But their faces are completely expressionless, and their bodies completely alien to ours which makes it hard for us to empathize or emote them unlike other creatures such as dogs or monkeys. Yet, multiple studies have indicated that an insect can learn abstract concepts and is capable of social behavior which is exhilarating.
The desire to map the network of a human brain is inspired by the notion that delineation and analysis of its architecture shall throw some light upon its complex working and its disorders following the idea that “structure drives behavior.” This holds true from the cellular level, where the entire set of proteins expressed by a genome, cell or an organism referred to as the proteome drives the behavior of the tiny cell to the social level, where it determines whom, we are friends with.
This brings us to the fact that mapping whole brains is extremely strenuous and time-consuming. A brain is required to be sliced into hundreds or thousands of individual tissue samples, all of which are visualized under an electron microscope wherein the pieces are reconstructed to form an accurate portrait of the brain.
Therefore, mapping anything like the human brain cannot be anticipated soon, possibly not even in our lifetimes, even with contemporary technology. It took a decade to give a detailed reconstruction of the connectome of the fruit fly at the level of local brain regions and interconnecting fiber bundles, one can only imagine! And, and even so the work took 12 years to be completely curated, it took an entire day per neuron for imaging alone. (Neurons fire at immense rate in your head to create the phenomenon of the mind being blown away)
Additionally, Sporns and Hagmann, in 2005, simultaneously but independently, coined this term “connectome” described as “a comprehensive structural description of the network of elements and connections forming the human brain.” A subsequent study following the idea with data from just 5 human subjects mapping the posterior medial cortex suggested that these structural connections shape the brain’s functional topology (Fig. 2)

A. Schematic description of connectome reconstruction with network nodes (i.e., Brain regions) and edges (i.e., connections) derived from anatomical T1-weighted and diffusion-weighted MRI data, respectively.
B. Four graph metrics commonly used to assess the topological organization of a (brain) network, including: (1) degree, representing the number of connections of each node i; (2) clustering, which is a measure of the extent to which nodes in a network tend to cluster together (i.e., how many triangles any node j is part of); (3) path length, which reflects the average number of steps it takes to get from any node i to any node j in the network (as a measure of network efficiency); and (4) modularity, which measures the extent to which a network can be decomposed into modules (illustrated here module x and y).
C. Connectome reconstruction from neuroimaging data showing a “rich club” organization with brain hubs and interconnecting rich club connections in red, “feeder” connections between hubs and non-hubs in orange, and “local” connections between non-hubs in yellow.
Source: Created using BrainNet Viewer https://doi.org/10.1371/journal.pbio.3002043.g001
“What we learned about code for fruit flies will have implications for code for humans. That’s what we want to understand-how to write a program that leads to human brain network.” notes Bert Vogelstein, M.D, Professor of Oncology at John Hopkins University.
This study is a milestone to further advances in prominent machine learning architectures, revealing more computational principles and inspiring new artificial intelligence systems. Along with that, it shall help to elucidate the pathophysiology of brain disorders such as explicating white matter tracts activated by deep brain stimulation (DBS) may be valuable in providing information about the neural circuit substrates (population of neurons interconnected by synapses) in mitigating treatment-resistant depression.
Looking at the minds of such organisms-smaller and simpler, helps us understand our own minds-larger and complex. Our experiences are that of a conscious entity constituting of neural mechanisms. And this brings us to question the fact that we might not be the only mindful entities on this planet. (Neurons form mindful connections to form a question mark)
References
- Winding, M., Pedigo, B. D., Barnes, C. L., Patsolic, H. G., Park, Y., Kazimiers, T., … & Zlatic, M. (2023). The connectome of an insect brain. Science, 379(6636), eadd9330. https://www.science.org/doi/10.1126/science.add9330
- Collin, G., & Whitfield-Gabrieli, S. (2023). Mapping the multimodal connectome: On the architects of brain network science. Plos Biology, 21(3), e3002043. https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3002043