Author: Manish Verma
Editor: Dr. Jitendra Kumar Sinha
The evolution of life on Earth has been shaped by the daily variation of light intensity and other environmental changes that result from the planet’s rotation. In response, a multitude of organisms have evolved biological clocks that synchronize their physiology, metabolism, and behavior to this 24-hour beat. As the sun rises and sets each day, our internal clocks adjust to ensure that we are awake and alert during the day and asleep at night. This process of synchronization is a complex area of research that explores how external cues influence our biological rhythms. Understanding the intricacies of circadian synchronization is crucial to maintaining our physical and mental well-being.
Although the mechanisms of these clocks differ across species, they all share the same basic functions: timekeeping, output, and entrainment.
To keep time, our biological clock requires a core oscillator and additional components that translate this oscillation into rhythmic biological activities. Additionally, the clock must remain synchronized with the environment and reset itself when necessary to maintain optimal health. This core oscillator is capable of being reconstructed in an in vitro environment and serves as a valuable tool for the investigation of the underlying molecular mechanisms that drive the processes of circadian timekeeping and entrainment.
To elucidate this concept, we shall direct our attention towards a minusucle aquatic unicellular microorganism known as cyanobacterium, specifically cyanobacterium Synechococcus elongatus PCC 7492. (Here, PCC 7492 is a specific strain often used as a model organism to study the molecular mechanisms of circadian rhythms and other physiological processes)
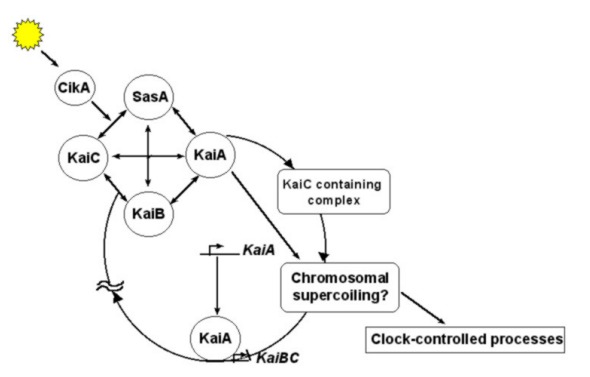
The circadian clock in Synechococcus elongatus relies on a core oscillator that progresses through its cycle via the interactions of KaiA, KaiB, and KaiC proteins. In the daytime, KaiA binds to an exposed peptide of KaiC, leading to KaiC autophosphorylation on two residues, S431 and T432. The KaiC complex attains a fully phosphorylated state near dusk, which promotes a conformational shift that facilitates KaiB binding. KaiB then acts antagonistically by capturing KaiA, thereby inhibiting further phosphorylation. The outcome of these interactions is a ~24-hour cycle of phosphorylation/dephosphorylation of the KaiC-based complex. Additionally, metabolic signals generated via photosynthesis serve as a substitute for sensory photoreceptors to synchronize the circadian clock.
The most noteworthy aspect of the KaiABC oscillator is that its phosphorylation/dephosphorylation cycle can be reconstituted and monitored using purified proteins in vitro.
Prior research has demonstrated that certain metabolic changes, specifically alterations in the ATP/ADP ratio and redox status of the quinone pool, that occur within cells during the transition to darkness are responsible for entraining the circadian clock. Modifying the ATP/ADP ratio or introducing oxidized quinone can lead to a phase shift in the phosphorylation cycle of the core oscillator in vitro.
These metabolic signals play a pivotal role in resetting the clock of cyanobacteria. For instance, a drop in ATP level has been shown to shift the phase of the oscillator by changing the timing of the peak of KaiC phosphorylation.
However, the in vitro oscillator cannot account for gene expression patterns as the basic mixture lacks the output components that link the clock to genes.
Two years ago, a team of researchers built a functioning circadian clock in a test tube, known as the “In Vitro Clock”. This allowed them to investigate the clock’s rhythms, its interactions and analyze how they regulate gene expression during different times of the daily cycle.
Recently, a study has devised a novel method to monitor the synchronization of the circadian clock with environmental cues in real time, thus extending the existing foundation of research in the field. This advancement allowed the researchers to delve deeper into the clock’s previously unexplored internal functions, including the transmission of time-setting signals from its core oscillator to the expression of genes responsible for maintaining proper clock functionality.
To investigate how the circadian clock undergoes molecular level resetting, comparable to humans’ adaptation to time zone changes during travel, researchers turned to aquatic single-celled organism, cyanobacterium. Using the In Vitro Clock, they were able to study this organism’s clock, which shares similar functions with that of a human, and track results in real-time using a new high-throughput method. The most significant discovery they made was related to the circadian clock’s component responsible for relaying the rhythm from the oscillator to gene expression. They observed that the catalyzing enzymes called kinases, which rhythmically modify the regulator to generate circadian gene expression, also play a crucial role in the clock’s functioning.
According to the author, the primary research focus after the discovery of the circadian clock has been on the core oscillator for nearly two decades. However, the author’s recent findings suggest that the kinases, which were previously considered as output components, are crucial to the entire mechanism of the circadian clock.
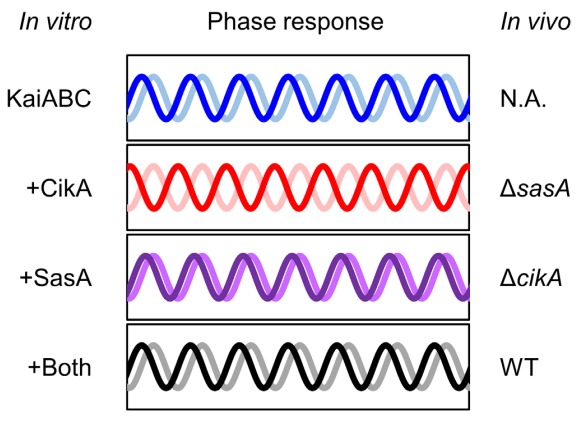
The discovery was compared by the author to the observer effect in physics, where the act of observation affects the observed system. The study found that the kinases, previously thought to be only output elements, are involved in the entire circadian clock mechanism, and disturb the core oscillator to obtain timing information.
According to the author, “kinases exert a physical influence on the core oscillator, enabling them to ascertain the time of the clock, and thus are integral components of the system.” The study revealed that two kinases are required for accurate functioning of the circadian clock, with the core oscillator being the “gears” of the clock and the kinases being the “hands” of the clock. The two kinases are responsible for stabilizing and perturbing the resetting signal, respectively, and are essential for proper timekeeping.
This research has therefore identified a critical aspect of the circadian clock’s internal mechanism, highlighting the significance of the interplay between the core oscillator and the kinases in regulating the body’s daily rhythms.
Keywords:
Circadian Rhythm; Biological Clock; cyanobacterium Synechococcus elongatus; KaiABC
References:
- Fang, M., Chavan, A. G., LiWang, A., & Golden, S. S. (2023). Synchronization of the circadian clock to the environment tracked in real time. Proceedings of the National Academy of Sciences of the United States of America, 120(13), e2221453120. https://doi.org/10.1073/pnas.2221453120
- Pandi-Perumal, S. R., Cardinali, D. P., Zaki, N. F. W., Karthikeyan, R., Spence, D. W., Reiter, R. J., & Brown, G. M. (2022). Timing is everything: Circadian rhythms and their role in the control of sleep. Frontiers in neuroendocrinology, 66, 100978. https://doi.org/10.1016/j.yfrne.2022.100978
- Reddy, S., Reddy, V., & Sharma, S. (2022). Physiology, Circadian Rhythm. In StatPearls. StatPearls Publishing.
- Sinha, J. K., Vashisth, K., & Ghosh, S. (2022). The importance of sleep studies in improving the health indices of a nation. Sleep medicine: X, 4, 100049. https://doi.org/10.1016/j.sleepx.2022.100049
- Sachdeva P, Ji S, Ghosh S, Ghosh S, Raghunath M, Kim H, Bhaskar R, Sinha JK, Han SS. Plausible Role of Stem Cell Types for Treating and Understanding the Pathophysiology of Depression. Pharmaceutics. 2023 Mar 2;15(3):814. doi: 10.3390/pharmaceutics15030814. PMID: 36986674; PMCID: PMC10058940.